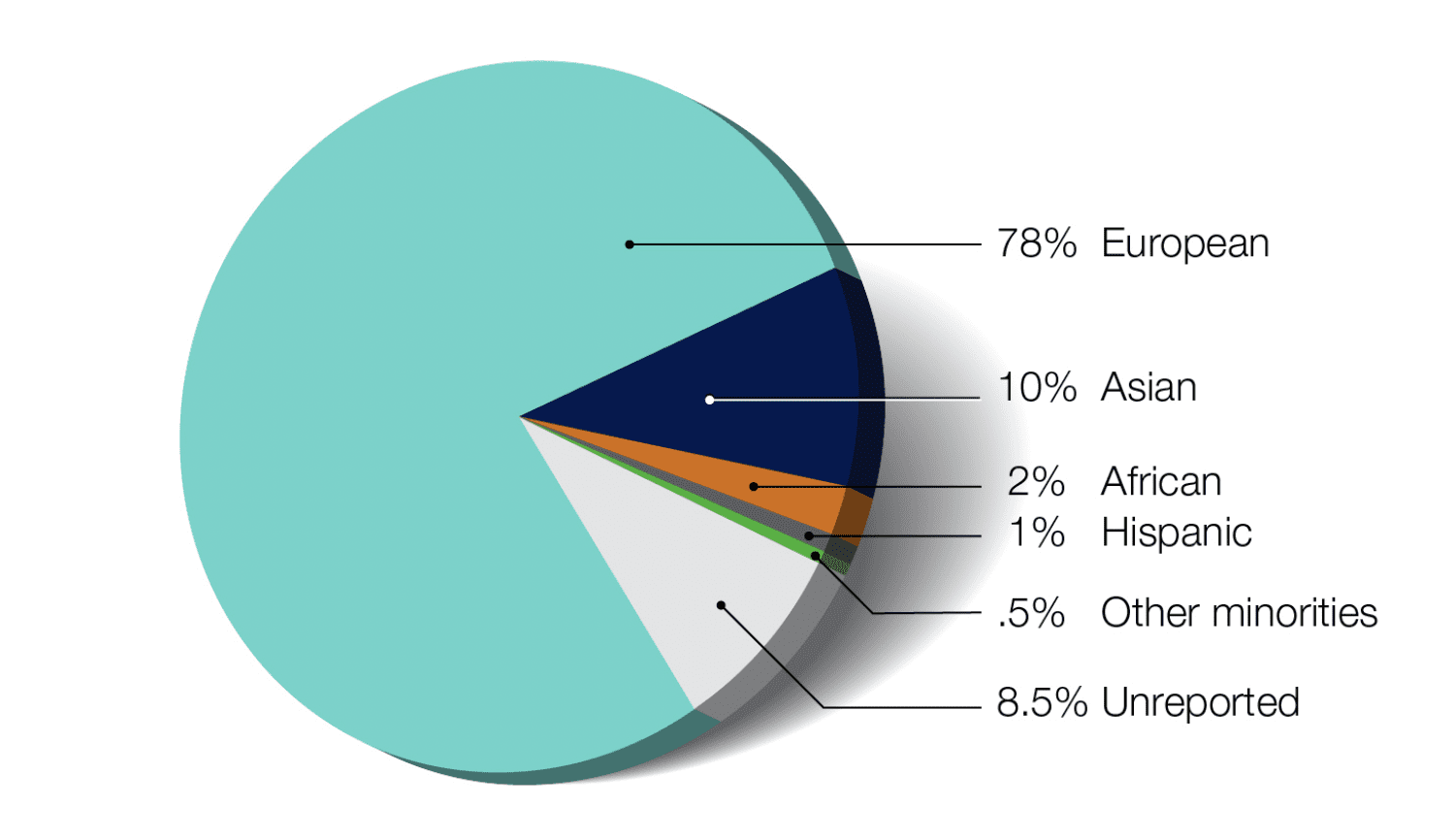
The First Version of SelfDecode
SelfDecode was first released in 2016 as the first step to understanding your genetics. It was an easy way to look at your genetic variants (SNPs) and find the different peer-reviewed studies attached to those SNPs.
But, there was still a lot of manual labor that needed to be done on the user side. Plus, we didn’t want people to mainly look at individual SNPs because we knew that it’s usually not predictive of conditions or traits that involve many variants (complex traits like mood, gut health, weight, etc…).
So in 2017, we started to release DNA Wellness Reports that contained up to 100 SNPs for a variety of topics.
I had hired genetic scientists from top universities to work on building extensive and accurate reports, and they were the best on the market at the time.
These scientists were taking the most impactful variants and putting them through a special algorithm we created to add them up and give a final prediction score.
Rebuilding SelfDecode From the Groundup: Polygenic Risk Scoring
In 2018, studies started to come out demonstrating that even a few hundred SNPs were not enough to give an accurate prediction score for complex traits. Often, thousands or even millions of variants were impacting a complex trait like mood or cardiovascular disease. These variants were having small but cumulative impacts, so it was really important to analyze ALL individual SNPs to get the best risk score.
In that same year, the American Heart Association named polygenic risk scores as one of the major breakthroughs in research into heart disease and stroke.
While SelfDecode was more advanced than what was out there at the time, I realized it was lacking compared to the new methods coming out of academia.
The newest techniques involved what’s called Polygenic Risk Scoring, a term that has grown in popularity since then and will continue to grow well into the future.
I recruited a Chief Science Officer, Dr. Puya Yazdi, an expert in Polygenic Risk Scoring that could lead a cross-disciplinary team of scientists.
He explained to me that the predictive potential of what we were doing was quite limited. We looked at what every other company was doing and it turned out that their methods were similar or worse.
Predictive Power With Individual or a Handful of Variants
For any given topic, there are multiple studies published, and typically there’s no overlap in the SNPs that are significantly associated with a trait.
If you read up a lot in the area, you’ll see that studies usually can’t be replicated, and the impact for a given SNP is usually very small. The only way to get around this is through cutting-edge Polygenic Risk Scoring methods.
Even with well-known variants in MTHFR, the vast majority of studies on these SNPs can’t be replicated.
That’s why we set out to create a cutting-edge system that would outperform the prediction models that other companies or academia were capable of doing.
In 2021, we released the world’s first Polygenic Risk Scoring algorithm that looked at millions of variants per a topic – directly to the consumer market.
We initially called this “SelfDecode 2.0” to let people know that we upgraded the backend of our software, but honestly, I don’t think that did it justice. The upgrade was MASSIVE.
We went from being able to analyze a hundred SNPs per health topic, to now analyzing millions of variants per topic, which meant the most accurate risk scores, and the best personalized recommendations based on your DNA.
In fact, to this day, SelfDecode is still the only company using cutting-edge Polygenic Risk Scoring for the direct-to-consumer market.
Not only that, but we compared our methods to the leading academic and industry models and ours outperformed these. We are in the process of submitting papers that demonstrate our results.
How Do Polygenic Risk Scores Compare to Standard Methods?
One of the first things we set out to test was how much better the new methods were than the old methods.
Using public data sets, we gathered the data of 800,000 people that included their genetic information, along with their likelihood of getting various conditions or traits. This allowed us to test the predictiveness of different methods.
We looked at what happens when we put in individual and groups of SNPs (this is the method other companies use to produce their DNA health reports).
In most cases, it turned out that inputting groups of SNPs like this was not more predictive than a coin flip. However, our Polygenic Risk Predictions were typically having quite significant predictive power, and in genetics, that makes all the difference in the world.
It’s really important to realize this because genetics is unlike other industries.
If you buy a cheap Toyota, it may not be as fast or look as good as a Mercedes, but it performs all of the basic functions of a Mercedes. They are both vehicles that get you from point A to point B.
However, when it comes to genetics, the difference between 0 and 1 can mean the difference between something that is no better than a random number generator versus something that actually has good predictive power.
When it comes to your most valuable possession, your health, you don’t want to leave it up to a random number generator.
Good Tech is Not Cheap
When we initially released these pipelines for polygenic risk scoring, it was costing us around $25 per user to analyze the results. This compared to less than one cent before.
We put in a lot of effort to reduce the cost, but it still costs quite a lot to run these systems, even on a marginal basis.
Our bet is that consumers will choose quality over marketing hype when it comes to their health and go for the premium products.
This is one of the reasons we have a subscription model – because every year our algorithms and methods update and it costs us money to regenerate everyone’s reports annually. But we do this for people on the lifetime and annual plans.
Polygenic Risk Scores Are Not Absolute
Keep in mind, however, that polygenic scores do not provide an absolute risk for a disease.
For example, consider two people with high polygenic risk scores for having coronary heart disease. The first person is 22 years old, while the other is 98. Even if they have the same polygenic risk score or genetic predisposition, they will have very different risks of the disease over the next 5 years.
Your gene expression also changes through your lifestyle, diet, and supplement choices, so all of these have a huge impact on whether or not you develop the risk of a specific condition.
I really like seeing my predispositions, because it allows me to look at my personalized recommendations for ways to reduce the risk of those conditions. When I see I have a predisposition for a condition that is over the 90th percentile, I especially pay attention.
Other Companies Introducing Polygenic Risk Scores
We realized that there was a big problem of quality in the personalized health industry that stemmed from a lack of ready technology that can be offered at scale.
So we decided to start a business unit under our parent company called OmicsEdge that licenses these technologies and reports to other companies, so that we can increase the quality of analysis in the whole industry.
To date, there are 20 companies that will be building apps using our cutting-edge Polygenic Risk Scoring.
When Are Individual Variants Useful to Look At?
Sometimes, when forming a hypothesis about which recommendations can work better for you, it can help to look at individual or a group of variants.
I typically look at individual variants when I am looking at clues for a mechanism increasing my risk.
For example, it was important to see that my tryptophan hydroxylase gene was not very effective at creating 5HTP, and resulted in creating less serotonin. It makes sense why 5HTP is the single most helpful mood booster for me.
Importance of Ancestry Adjusted Polygenic Risk Scoring
According to the NIH, 78% of genomic studies are done on Europeans [1].
This is an important issue because when you’re dealing with a US audience, which is diverse, Polygenic Risk Score should take into account the ancestry in order to get the best results, and adjust the scores accordingly [1].
This is one of the issues preventing Polygenic Risk Scoring from entering the mainstream healthcare system, but is something we utilize at SelfDecode, as opposed to the other companies in the industry.
Why is SelfDecode The Only Company Doing Ancestry Adjusted Polygenic Risk Scoring?
The reason why we are the only ones on the market who have this technology is because it’s really hard to build and to do it at an industrial scale.
We had to build quite a large R&D team to put it together.
The only company that comes close to doing this is 23andme, but they aren’t doing it at the same scale as us. I haven’t seen any of their reporting looking at more than single-digit thousands of variants.
In academia, they typically build a model to test it out, but it doesn’t need to go into actual production and be used at scale.
It’s the difference between building a prototype of a product vs building it at scale. Building a prototype is always way easier than building something at scale.
So, to answer the question… Why did we spend $10M to rebuild SelfDeocde?
Because the world needed it and no one else was doing it. Because your health deserves the best science has to offer. And because we believe in accessible, precision health that has the ability to change lives.